| Student: | D.E. Payares Garcia |
|---|---|
| Timeline: | September 2021 - 1 September 2025 |
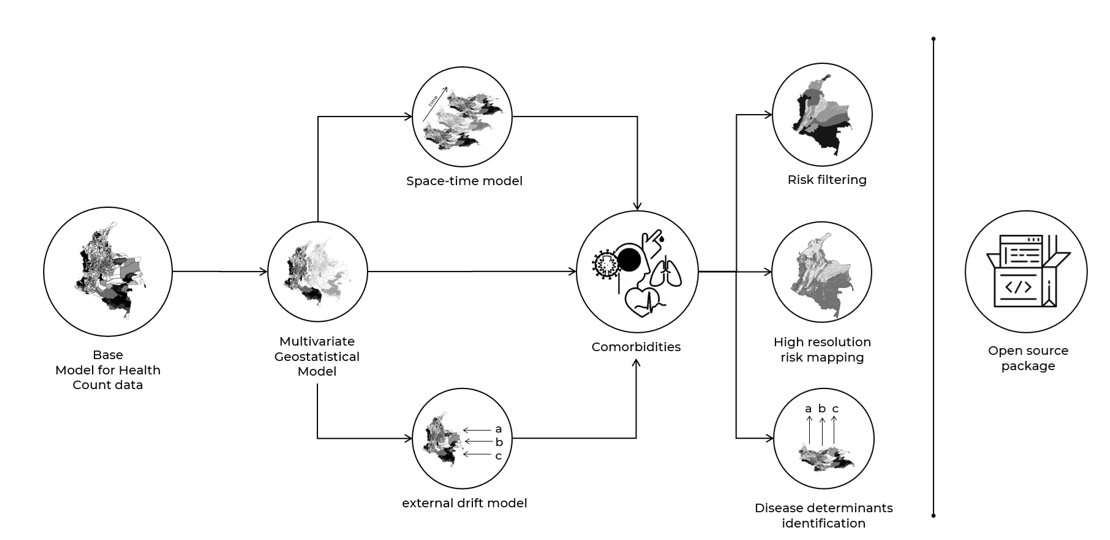
Comorbidity is a commonly observed pattern in epidemiology. As diseases rarely occur in isolation, more than one disease can affect a population simultaneously. From a spatial perspective, studying comorbidities can measure the stimuli diseases exert in each other's development and propagation. Furthermore, it can unveil interdependent mechanisms that obscure the real relationships between illness and risk factors. For example, Dengue prevalence cannot be only explained by predisposing factors (e.g. humidity, closeness to water bodies, poverty), as underlying comorbidities such as diabetes and pulmonary diseases also exacerbate it. This research proposes an adaptation of cokriging for health count data to understand the spatial process behind disease comorbidities. We will associate a multivariate geostatistical model with the Poisson distribution to model diseases' rate spatial variation. We will also introduce spatio-temporal and external drift adaptations of the model as both time and risk factors drive the epidemiological profile of diseases.
This work focuses on three main applications: i) risk filtering, ii) high-resolution disease mapping and iii) shared and non-shared disease determinants identification. This novel approach will be applied and tested with multiple diseases and datasets. This work also seeks to enhance the use of spatial statistical methods for non-statisticians and highlight the importance of open-source algorithms in medical research. By elucidating the spatial patterns of disease comorbidities and their potential influencing factors, this project aims to inform comprehensive and integrated public health policies and interventions, ultimately contributing to the mitigation of disease burden and improved population health outcomes.